Study aimed at improving brewing efficiency provides unexpected insights on evolution

Researchers at the University of Wisconsin–Madison made an unexpected discovery while studying a close relative of the yeast commonly used to ferment beer. The yeast left half its genetic material behind while evolving in the lab.
While the discovery could have benefits for applications like biofuel production and other industries, in this case it was a practical problem – how to improve beer by fermenting all the available sugars – that led to a discovery about biological adaptation.
“That reverses the paradigm where we often talk about fundamental curiosity-driven research enabling translation and applications,” said genetics professor Chris Todd Hittinger, an investigator with the Great Lakes Bioenergy Research Center and co-author of the paper published this week in the journal PLoS Biology.
The study started with an investigation of a strain of Saccharomyces eubayanus, a wild parent of the hybrid yeast strains used in lager-style beer fermentation.
A key trait of yeasts used in beer production is the ability to break down maltose, the most abundant sugar in the material used to brew beer. The team chose a yeast strain that could not metabolize maltose, despite having many of the genes required.
In a lab experiment, researchers grew the yeast on maltose, selecting descendants that thrived. Eventually the yeast regained the ability to eat the malt sugar.
However, it was not immediately clear how the yeast changed.

“We kind of banged our heads against the wall because we couldn’t really make sense of figuring out the underlying genetic changes of these adaptively evolved isolates that had improved their maltose metabolism,” said John Crandall, first author of the paper and a doctoral student in Hittinger’s lab in the Wisconsin Energy Institute.
There was not just a simple mutation in any specific gene — the change was much more dramatic: the yeast had lost half of its genetic material during the experiment. More specifically, all chromosomes lost their pair, turning from diploids (with two copies of each chromosome, like humans have) to haploids (with a single copy of each, like our reproductive cells).
The number of sets of chromosomes in organisms, known as ploidy, is one of the most fundamental defining aspects of organismal biology.
“It defines the structure and content of the genome, which then has all sorts of implications for life cycle and evolutionary trajectories,” Crandall said.
Ploidy changes in yeasts have been documented before, but usually haploids become diploids and not the other way around. The ploidy change in yeasts also frequently causes a change in cell type, which directly affects sexual reproduction. But here, it was affecting sugar metabolism.
“So there were lots of different factors to sort of disentangle between the absolute ploidy, the cell type, how those different things contributed to fitness [growth and reproduction]. And that interested us, because there are some different theoretical frameworks about why absolute ploidy itself might impact fitness,” Crandall said. “But there wasn’t a good model for why cell type should affect fitness in this condition, which is also why it’s particularly surprising that it did.”
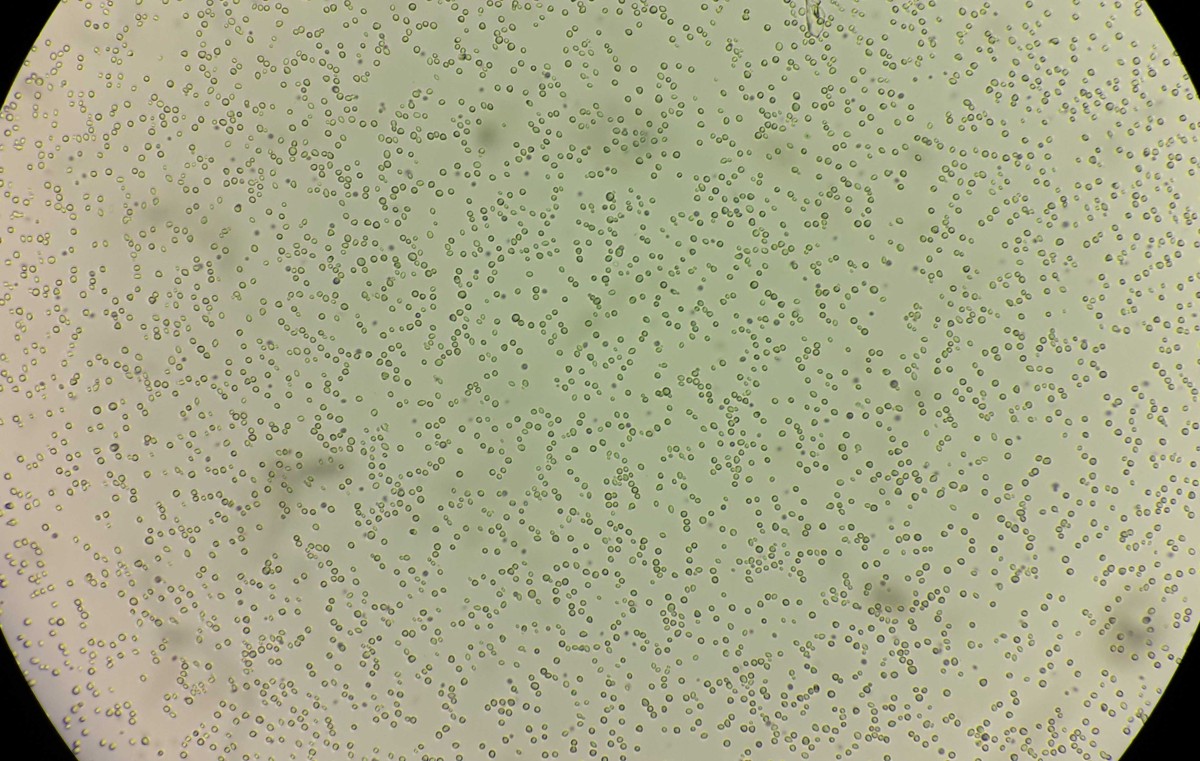
The new study is perhaps the clearest mechanistic insight into a ploidy change that directly affects growth and metabolism, which could help uncover better microbes used in medicine, brewing, biofuel production and other industrial uses.
When evaluating microbes for useful traits, researchers often have only one strain – or individual – to represent the entire species, even though there’s often great diversity within a microbial species.
Crandall said looking at different ploidy states could reveal even more desirable characteristics.
“When trying to improve a desirable trait in a strain, we wouldn't typically think to start messing with its ploidy or cell type,” Crandall said. “But we're suggesting that people could do just that, since it might unexpectedly influence that desirable trait.”
This research was supported by grants from the National Science Foundation (DGE-1747503, DEB-1442148 and DEB-2110403) U.S. Department of Agriculture (Hatch Projects 1020204 and 7005101), Department of Energy (DE–SC0018409 and DE-AC02-05CH11231) and National Institutes of Health (5T32GM007133).