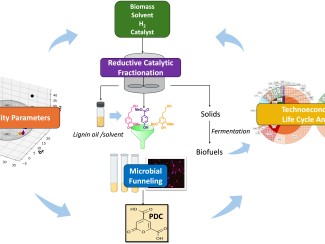
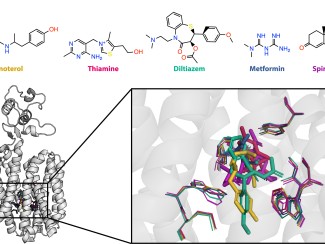
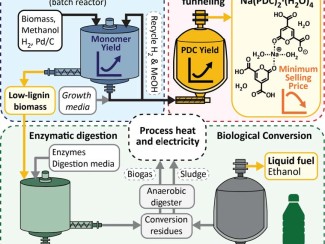
Study investigates whether yeasts’ preferences for certain genetic arrangements can predict their ability to ferment xylose, the second-most abundant sugar in plant biomass.
The Science
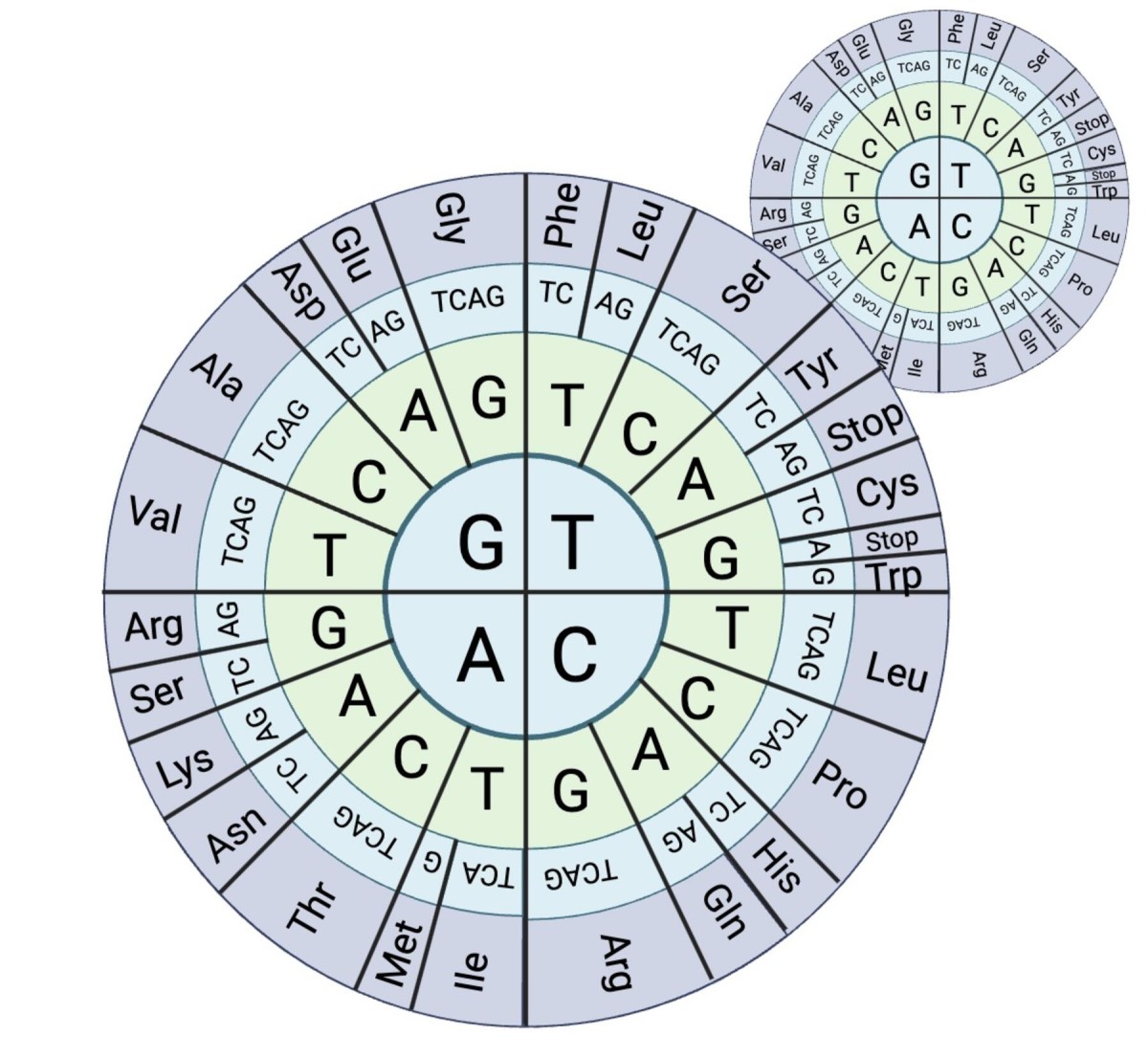
Xylose is the second-most abundant sugar in plant biomass, but most yeasts cannot eat xylose even though many have the required genetic pathway. For this study, researchers looked at codons, the building blocks of DNA, in an attempt to predict which yeasts would be able to consume xylose. Codons group into three to code for one amino acid. The key to codon optimization is that different codon “triplets” can code for the same amino acid. The graphic above demonstrates this principle. The direction of the genetic code from the second to the third base, or ring, shows that a codon triplet has the option of which codon to pair with for a given amino acid. The species then chooses which codon “recipe” is easiest to read for the amino acid needed for xylose metabolism. The study showed that codon optimization can help predict whether a species can consume xylose.
The Impact
Because xylose is so abundant, this research is important for the production of plant-based replacements for fuels and other products currently derived from petroleum, a critical step in slowing climate change. Though many of the yeasts in the ancient group being examined are unable to ferment xylose, the vast majority contain the necessary genetic code to consume xylose. Using this predictive power of codon optimization, scientists could insert genes needed to ferment xylose into existing industrial strains, enabling them to ferment all the available sugars in biomass.
Summary
This study examined 332 yeast species. By submerging them in a xylose-rich medium, researchers were able to observe which species consumed xylose.
The results showed the baseline requirement for xylose fermentation is a complete XYL genetic pathway. This XYL pathway is the enzymatic pathway required for xylose consumption, and it contains three primary genes: XYL1, XYL2, and XYL3. The number of copies of XYL1 was highly correlated with the yeast’s ability to consume xylose, but this was not the case for XYL2 or XYL3. Instead, codon optimization of XYL3 was significantly correlated with the yeast’s ability to consume xylose, while codon optimization of XYL2 was significantly correlated with the yeast’s growth rate on xylose. Overall, yeasts with multiple copies of XYL1 and highly optimized XYL2 and XYL3 genes can help to predict whether the yeast species will consume xylose.
These indicators will help identify xylose-fermenting yeasts from genome sequence alone, unlocking the potential of a relatively untapped source of biofuels and bio products.