In this series, we learn more about what inspired our talented graduate students and postdoctoral researchers, what brought them to their field of study, and the questions that drive their work at the Wisconsin Energy Institute (WEI).
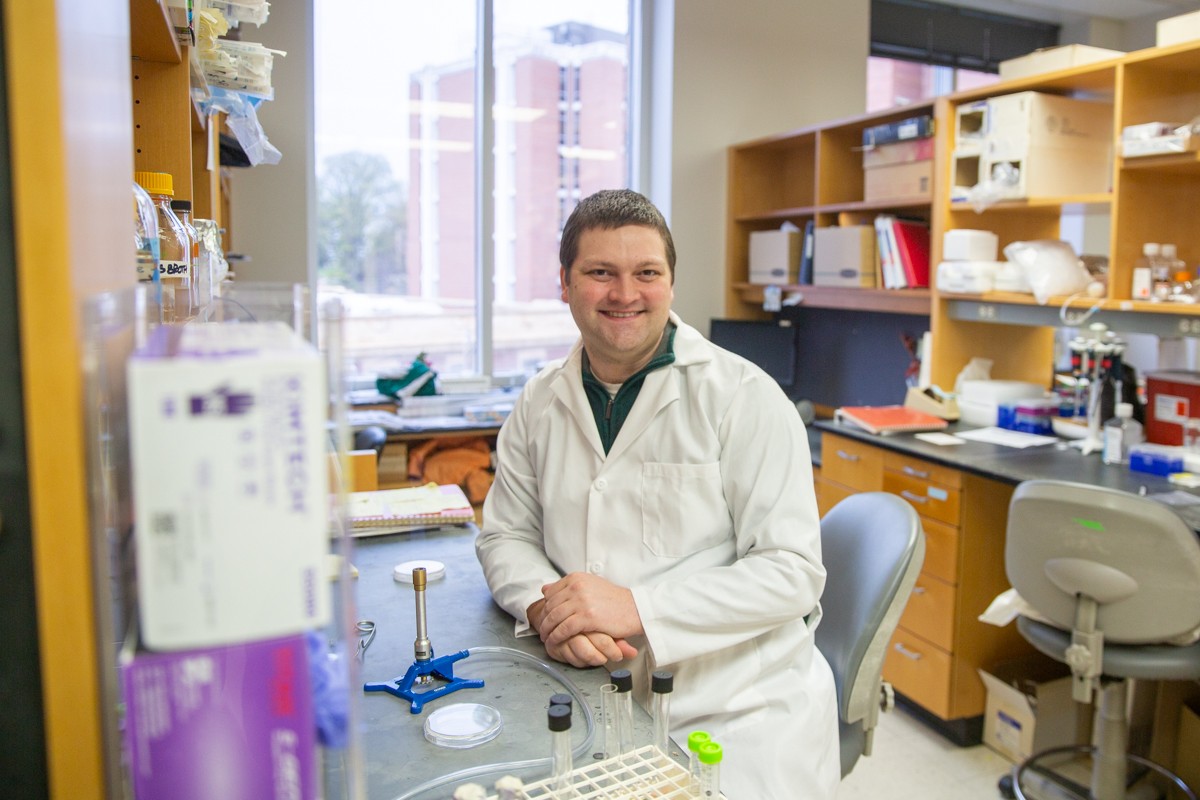
Today we spoke with Daniel Parrell, a postdoctoral research associate in the Wright Lab working with cryo-electron microscopy and cryo-electron tomography. He received his undergraduate degree in microbiology from the University of Wisconsin–Madison and his PhD in microbiology and molecular genetics at Michigan State University (MSU). He provides insight on his work and what it’s like to be a postdoctoral researcher.

Tell me about your background:
I'm a post-doctoral researcher in Elizabeth Wright's group. I have been here for four years now. I received an NIH (National Institutes of Health) fellowship in 2021 to support our research. I received my PhD at Michigan State University in microbiology and molecular genetics. I received an undergraduate degree here at UW–Madison in microbiology. My parents and family are from the Black Earth area, which is just outside Madison.
How did you begin working with the GLBRC?
During my PhD, I gained a lot of skills with microbiology and molecular genetics working with sporulation (formation of spores) in a bacterium named Bacillus subtilis. Coming to Madison for a postdoc, I was looking to learn a new technique. Cryo-electron microscopy and cryo-electron tomography were just arriving on campus, which is what Dr. Wright's lab specializes in. And I thought that my skill set could help advance some of the projects that work with bacteria in her lab.
What is cryo-electron microscopy?
It's electron microscopy but done at cryogenic (very low) temperatures. So, during transmission electron microscopy, which we can think of as analogous to light microscopy, we're sending electrons toward your sample and you can see what is on the stage based on your sample essentially blocking, or scattering, some of those electrons. This creates a projection, or image, of your cell and the things inside. With light microscopy, you are fairly limited in resolution when it comes to bacteria. However, with electrons, you can get a much higher resolution, because of the much shorter wavelength of electrons.
The cryo part is one of the really interesting advances that has expanded in the last 10 to 15 years. If you can keep your sample frozen in the microscope, you can actually look at your sample frozen in what we call the “native state.” Often with electron microscopy, you have to use what are called negative stains, many containing heavy metals, in order to see your sample. But recently, the microscopy companies have developed cameras, called direct electron detectors for their systems that work a lot better than previous generations of cameras.
Essentially these cameras can detect electrons as they reach the camera and they have fast frame rates that allow us to “de-blur” the images, in a process called motion correction. With these detectors, we can see samples that haven't been treated with stains much better than before. Cryo-electron microscopy is a really powerful tool where we can solve the structures of proteins, and look at the details inside of human and bacterial cells that we haven't been able to before.
Cryo-electron tomography is kind of like a CT scan. Essentially, we take several pictures of the same cell at different angles. You can imagine an arc of pictures that are projections of the cell as we tilt the stage. We then use those pictures to reconstruct the cell in three dimensions into what we call a tomogram. You can imagine going slice by slice through this volume of the cell and seeing the structures present in the cell as you move your way through. From here, we can model the structures inside the cell.
What sparked your passion for research?
I've always wanted to be a microbiologist, I would say, from early on in my education. I worked in labs as an undergraduate, and I was always interested in how proteins and really small things worked. And with bacteria you can study those things quite easily and rapidly. Within microbiology, you also have all of the different subdivisions of biology: you have ecologists, you have protein biochemists, you have geneticists, and you have people who are interested in pathogenesis and disease-causing organisms. So it's kind of a community unto itself of all different types of biologists, which makes for such a fun and diverse community. That's really what drew me to like microbiology, and then working with the GLBRC it has been nice to take that interest and apply it to something that's going to help affect the world.
Working with the GLBRC it has been nice to take that interest and apply it to something that's going to help affect the world.
Daniel Parrell
Are you working on any projects?
Yes, we have a project where we were doing experiments with an antibiotic called chloramphenicol, which stops bacteria from making new proteins. We were trying to see an effect with the nucleoid, or the DNA, after treatment. We saw an effect with light microscopy, and when we went to use cryo-EM we were surprised to see changes to these storage granules for polyhydroxybutyrate (PHB). We saw them grow really big in size. And so it has been kind of a little tangent that we've gone on trying to understand what's going on inside the cell to create this additional storage of essential carbon. PHB is where cells shuttle excess carbon, and we use the power of the new electron microscopes and their new detectors to take what we call tomograms of these cells. And we use those models to estimate how much PHB the cell is accumulating after our antibiotic treatment.
I worked with Dr. Rachelle Lemke in Dr. Tim Donohue’s lab to verify the PHB accumulation with gas chromatography-mass spectrometry in the Wisconsin Energy Institute (WEI) building. Essentially we were purifying this bioplastic from the cell and quantifying it. We were trying to verify whether measurements from our models match with what we can extract in the lab, and it actually worked really well. Thinking more broadly, understanding that cells are accumulating PHB under these conditions might help us understand what kind of processes to try to perturb going forward in order to better accumulate this bioplastic in the cell.
Describe your typical work week:
Work in the lab can be varied, which is part of the fun of being a researcher. I'm actually writing a paper currently so some of my time is spent at the desk. When I am doing lab work I might be growing bacterial cultures and collecting samples that I can bring over to WEI for GC mass spec. I also purify proteins for use with other experiments, and I will freeze either of those samples for electron microscopy work.
I also spend some time at the computer, working with images, making tomograms and the 3D models that I was talking about earlier. It is a fun challenge to find those nice looking, and representative, images from your work as you write up your paper. It is also a joy to spend some time reading and digging into the literature to make sure you are citing everything in the field that leads up to your own work. And of course working together with collaborators to revise the writing is a fun way to bring everyone’s great ideas together.

What’s something you’re proud of?
I would say that the interactions that I'm most proud of are with the students that I've worked with at Michigan State and at UW–Madison. I've worked with several undergrads now. And most of them have gone on to do really great things. Some have gone to medical school, dental school, or are working at pharmaceutical companies, a few have gone to graduate school, and I think one of them even has her PhD now, which is kind of weird because I feel like I just got mine. I enjoy seeing them succeed and do such great things in and out of the lab. Seeing them learn and have the little aha moments. For example with something like polymerase chain reaction. They learn about DNA polymerases in class, but now they're actually using that enzyme to amplify DNA for experiments. It's fun to see them put things like that together and be like, “Oh, that's what we're doing.”

How does it feel to be back in Madison?
I’m really enjoying it. One of the difficulties with jobs in an academic setting is that you're often limited geographically for where you can work. So I feel really fortunate that I'm able to be here, near to my family, to have their support, because I have a lot of colleagues that are postdocs who don't have that luxury. I'm thankful for the opportunity to work here, and for the mentorship I got from Dr. Wright and Dr. Donohue.
What do you like to do outside of work? I like to play soccer with friends of mine. However, in December, I tore an ACL while skiing, so I've been on the sideline for a bit. I also like long-distance running and I do some amateur photography. I also have a solid group of friends in Madison that I enjoy seeing, and of course, I love being with my family. Having those connections close by is important to me.