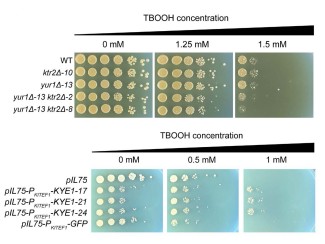
Some microbes are skilled at turning plants into biofuels and useful chemicals. Persuade them to do this efficiently enough, and you have the foundation of a bioenergy refinery that generates fuels and chemicals from renewable sources rather than fossil fuels.
Researchers can improve microbes’ productivity through precise genetic modification with CRISPR/Cas9, the gene editing tool that recently earned its inventors a Nobel prize. Now, Great Lakes Bioenergy Research Center (GLBRC) scientists have created an easy-to-use web tool called “CRISpy-pop” to streamline the process. CRISpy-pop allows users to refer to the genomic data of an entire population of biofuel-producing microbes to determine the best places to splice its DNA.
To make sure Cas9—the protein that does the dirty work—cuts in the right spot, scientists design single guide RNA (sgRNA) as a chaperone for the gene-splicing dance. Use the wrong guide, and you may not get the steps you want.
GLBRC talked with Hayley Stoneman, former undergraduate researcher in the lab of Chris Todd Hittinger and lead author on a new publication, about the science behind CRISpy-pop. Stoneman cut to the chase of this advance to explain how CRISpy-pop takes the guesswork out of choosing the best guide RNA to lead your desired modification steps.

What major challenge is CRISpy-pop looking to address?
For a guide RNA to be effective, it should have a high probability of targeting the intended DNA region without having any off-target, or unintended, effects to the genome. So this application is meant to ease that design step in choosing guide RNA sequences.
The special part is that it incorporates all of the sequences from the 1002 Yeast Genomes Project [an effort that genetically sequenced 1011 strains of the model bioenergy chassis Saccharomyces cerevisiae], as well as other GLBRC-designed strains. This gives the user more control over population-level diversity.
Why is it important to look at population data – the differences in strains of the same yeasts?
The level of diversity within the yeast genome is an order of magnitude greater than our own genome. An individual strain can be edited, but if you want to expand a study to an entire population, then it's really important to understand the diversity between strains. A single nucleotide difference in the targeted sequence can cause the guide RNA to malfunction and the Cas9 protein to be unable to attach to the sequence you want to alter. That’s why diversity is really important to consider when doing population-level studies.
Why would somebody want to be able to edit the whole population as opposed to just a single strain?
Different strains have different properties that affect tolerances to industrial stresses and their potential for specific biofuels and bioproducts. If you were designing a genetic modification that you wanted to be able to use in every S. cerevisiae strain in the future, you would want to be able to apply it to as many strains as possible. Otherwise, there's a good chance that it wouldn't be able to work and you'd have to design a whole different system.
How does CRISpy-pop make this process easier for bioenergy researchers?
All a scientist has to do is enter the genetic target – a gene or a specific sequence – and CRISpy-pop spits out all the possible guide RNAs that could work with the target they’re looking to alter. Because it has this nice GUI (graphical user interface), users can filter and look for ones in a specific location. So basically, it gives anyone working on this all the options. It’s also super useful because there are no other tools that incorporate this many strains. Plus, it's very, very easy to use.
Who else could benefit from using this kind of tool?
Anybody who's doing CRISPR-driven genetic modification in S. cerevisiae, and especially those who are doing them on a larger population scale, will benefit from using this program. It also has support for Zymomonas mobilis, another commonly used bacteria for bioenergy research, as well as a few other yeast species. The app also has a target custom sequence feature so any researcher can just copy and paste a sequence from any organism and it'll work.
GLBRC researchers in the Hittinger lab and elsewhere have already begun using CRIPy-pop to modify S. cerevisiae to more efficiently produce isobutanol and other chemicals and products. Try it out here.