A large evolutionary analysis reveals protein features linked to the ability to break down a wide range of complex sugars.
Turning bioenergy crops into fuels and other products requires breaking down the complex mixture of polysaccharides found in plant material. Glycoside hydrolase family 5 (GH5) is a large and diverse family of enzymes able to digest a wide range of polysaccharides. In a new study led by Evan Glasgow and Kirk Vander Meulen, researchers at the Great Lakes Bioenergy Research Center (GLBRC) described the functional diversity of members of a GH5 subfamily to explore the structural origins of their broad substrate specificity, a step toward engineering better enzymes for converting biomass into biofuels and other specialty bioproducts.
Broad-specificity enzymes offer the potential to simplify the complex enzyme mixtures used to produce fermentable sugars from biomass. The identification and analysis of a group of glycoside hydrolases able to break down multiple substrates with high efficiency offers clues to the sequence and structural features related to broad specificity, which will help researchers design cheaper and more efficient enzyme mixtures for industrial applications.
In the new study, the researchers expressed the catalytic domains of 243 GH5 members, including 238 in the GH5_4 subfamily, and quantified their activity against multiple polysaccharide substrates: lichenan, xylan, beta-1,4-mannan, and xyloglucan. A phylogenetic analysis based on the measured enzymatic activity identified three major GH5_4 clades with distinct activity patterns. Enzymes in Clade 3 showed a high level of activity against both lichenan and xylan, about one order of magnitude higher than the averages in Clades 1 and 2. Interestingly, lichenase and xylanase activity appeared to be strongly correlated across the subfamily, with a weaker positive relationship noted between lichenase and xyloglucanase activity.
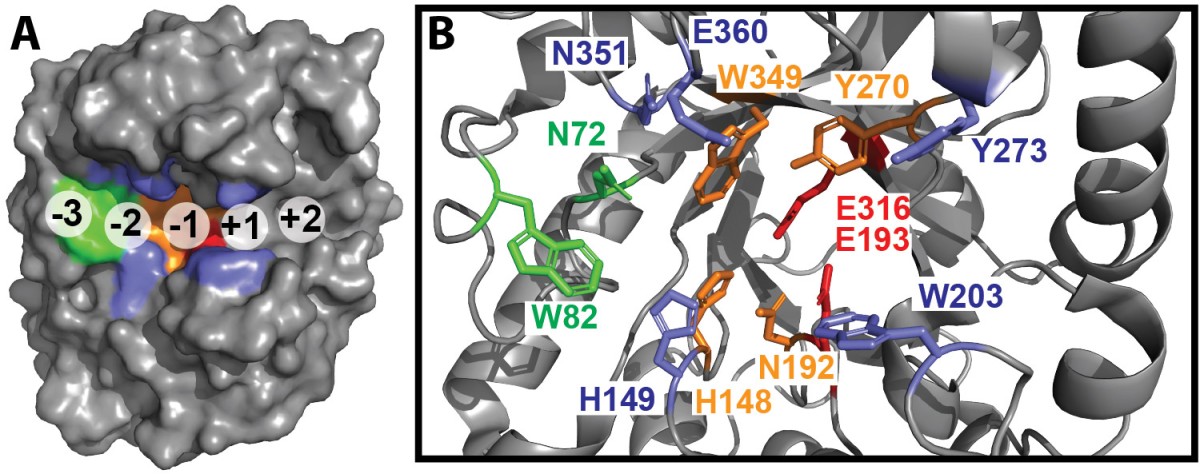
The team used the structure of CelE, a previously characterized Clade 3 enzyme with activity against all four substrates, to identify structural elements associated with this broad enzymatic activity. They identified two variably conserved residues in the binding cleft (H149 and W203) linked to strong activity across the substrates.
The findings show that activity against multiple polysaccharide substrates is relatively common among GH5_4 enzymes, and that it may be possible to use sequences to predict and design enzyme activity against different types of biomass.