Researchers from the University of Wisconsin–Madison and Vanderbilt University will use a $2.2 million grant from the National Science Foundation (NSF) to chart the evolution of over one-thousand budding yeast species across the span of four hundred million years.
The ambitious research project, led by UW–Madison genetics professor Chris Todd Hittinger and Vanderbilt biological sciences professor Antonis Rokas, will study how metabolism evolved in all known species of budding yeasts. By characterizing each species’ genotype – its collection of genes – and phenotype – its metabolic traits – the research team can visualize the relationship between genes and metabolism in a genotype-phenotype (G-P) map. The G-P map will help determine how varying genotypes across more than one-thousand yeast species has led to different phenotypes, which is a major question in the field of biology.
Understanding how phenotypic diversity is encoded in genomes and how it changes during the course of evolution can tell us a great deal about how life works.
Chris Hittinger
This five-year grant builds upon the pair’s previous Dimensions of Biodiversity grant from NSF, which sequenced the genomes of all known species of budding yeasts. Hittinger, Rokas, and their students and trainees will investigate the rules governing the evolution of the G-P map using the metabolisms of budding yeast species, which are as genetically diverse as the plant and animal kingdoms.
Budding yeasts are named for their rapid and unique cell division, in which smaller daughter cells will “bud” off a mother cell in as rapidly as an hour, in some conditions. These organisms are a good model for studying the evolution of the G-P map because their metabolisms allow for predictions about biochemical traits directly from genomic data. Since the genomes of almost all budding yeasts have already been sequenced, their metabolisms can be used to predict these traits more rapidly than other organisms.
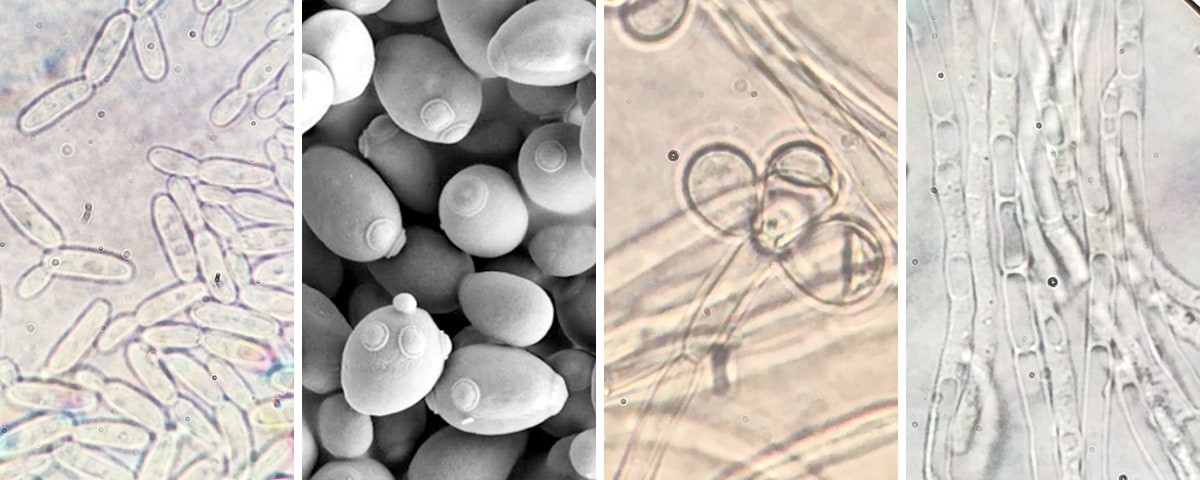
The project is funded through NSF’s Rules of Life program, which supports research seeking to investigate generality in biology. This process begins with a specific observation from a specific system or organism that can be widely applied across biological systems.
Defining the G-P map represents the first step towards harnessing budding yeasts’ carbon metabolisms for a sustainable bioeconomy. Various industries, including brewing, baking, and bioenergy, utilize budding yeasts’ carbon metabolisms for fermentation. Since this metabolism varies widely between each budding yeast species, it is vital to understand each species’ metabolic pathways to engineer them for biofuels and bioproducts.